class: center, middle, inverse, title-slide # Lecture B ## Exhaustive and non-exhaustive algorithms with resampling ### Samuel Müller and Garth Tarr --- ## Overview <br></br> B1. Cross validation B2. .blue[The `mplot` package] * .red[Artificial example]: *Information criteria can be blind* * .blue[Variable inclusion plots] * .blue[Model stability plots] * .blue[Bootstraping the lasso] B3. Subtractive stability measures --- class: segue # B1. Cross-validation for model selection --- ## Resampling procedures - .orange[Cross-validation (CV)] and .red[bootstrapping] are the two most popular classes of .blue[resampling methods] in model building - In model building, resampling is often used to - assess the <span style="color:blue">stability</span> of a model - infer on model parameters (tests, <span style="color:blue">confidence intervals</span>, point estimates, <span style="color:blue">bias</span>) - measure .blue[prediction error] --- ## Cross-validation algorithms - .orange[Cross-validation] schemes can be implemented in most statistical frameworks and for most estimation procedures - The key principle of CV is to .blue[split the data] into a .blue[training set] and a .red[validation set] - .blue[Estimators] are .blue[built on the training set] - .red[Validation set] is used .red[to] validate/.red[assess] the .red[performance] of the estimators, e.g. estimating their prediction risk - This training / validation splitting is repeated several times --- ## 10-fold cross validation <img src=imgs/k_fold_cv.jpg width=950 /> --- ## Cross-validation algorithms The most popular .orange[cross-validation schemes] are, - .blue[Hold-out CV], based on a single split of the data - <span style="color:red"> `\(\color{red}{k}\)`-fold CV</span>: the data is split into `\(k\)` subsamples. Each subsample is successively removed for validation, the remaining data being used for training - .blue[Leave-one-out] (LOOCV) which corresponds to <span style="color:red"> `\(n\)`-fold CV</span> - <span style="color:blue">Leave- `\(q\)`-out</span> every of the `\(\binom{n}{q}\)` possible subsets is removed for validation --- ## CV prediction error - In `\(k\)`-fold CV the data is split into `\(k\)` subsamples (the `\(k\)` folds) - Each subsample is successively removed for .red[validation], the remaining data being used for .blue[training] - Thus, for each of the `\(k\)` folds `\(f=1,\ldots,k\)`, `\(\iota_f\subset\{1,\ldots,n\}\)` denotes the `\(n_f = |\iota_f|\)` observations in the `\(f\)`th fold and we calculate `$$\text{CV}_{(k)} = \frac{1}{k} \sum_{f=1}^k \text{MSE}_f = \frac{1}{k}\sum_{f=1}^k \frac{1}{n_f}\sum_{i\in \iota_f} [y_i - \hat y_{\alpha i}(-\iota_f)]^2$$` - Similarly, for estimating classification errors --- ## Choice of `\(k\)` affects the quality of the CV error - If `\(\color{blue}{k=2}\)`: split-sample CV - CV error estimates .blue[biased] upwards because only half the data used in the training samples - If `\(\color{red}{k=n}\)`: LOOCV - CV estimates have .red[high variance] because in the above equation a total of `\(n\)` positively correlated quantities are averaged; - .blue[Standard choices] are `\(\color{blue}{k=10}\)` (default in `cv.glmnet` through option `nfolds=10`) or `\(k=5\)`: This balances bias and variance - The `\(k\)` partly overlapping training sets can be used to learn more about the stability of selected models --- class: segue # Case study: selecting `\(\lambda\)` for Lasso --- ## How to best choose `\(\lambda\)`? - Many options! - .blue[Use information criteria along the lasso], ridge, or elastic-net .blue[path] - Use cross-validation - as an alternative to `lambda.min` often `lambda.1se` does give better performance in terms of prediction - `lambda.1se` is the largest `\(\lambda\)` at which the MSE is within one standard error of the minimal MSE (at `lambda.min`) --- class: eg ### Diabetes example ```r data("diabetes", package = "lars") library("glmnet") x = diabetes$x2 # includes squares and interactions y = diabetes$y lasso.fit = glmnet(x, y) length(lasso.fit$lambda) ``` ``` ## [1] 100 ``` ```r lasso.predict = predict(lasso.fit, newx = x) dim(lasso.predict) ``` ``` ## [1] 442 100 ``` --- class: eg ### Diabetes example: AIC to select `\(\lambda\)` ```r lasso.aic = rep(0, 100) sigma.hat = summary(lm(y ~ x))$s for (m in 1:100) { yhat = lasso.predict[, m] k = lasso.fit$df[m] lasso.aic[m] = sum((y - yhat)^2)/(sigma.hat^2) + 2*k } which.min(lasso.aic) ``` ``` ## [1] 32 ``` ```r lasso.fit$lambda[32] ``` ``` ## [1] 2.524812 ``` --- class: eg ### Diabetes example: CV to select `\(\lambda\)` ```r set.seed(2020) lasso.cv = cv.glmnet(x, y) plot(lasso.cv) abline(h = min(lasso.cv$cvm) + lasso.cv$cvsd[which.min(lasso.cv$cvm)], col = "blue") ``` <img src="lecB_files/figure-html/unnamed-chunk-3-1.png" width="864" /> ```r lasso.cv$lambda.1se ``` ``` ## [1] 5.314486 ``` --- class: eg ### Diabetes example: Predictors selected by .bold[.blue[AIC]]: ```r aic_betas = glmnet(x, y, lambda = lasso.fit$lambda[32])$beta as.matrix(aic_betas)[as.matrix(aic_betas)>0, ] ``` ``` ## bmi map ltg glu age^2 bmi^2 ## 500.775931 262.976223 469.918325 25.416346 17.558198 42.899478 ## glu^2 age:sex age:map age:ltg age:glu sex:map ## 76.518096 115.463280 30.647942 12.662471 9.365541 7.745166 ## bmi:map ## 90.571069 ``` Predictors selected by .bold[.red[CV]]: ```r cv_betas = glmnet(x, y, lambda = lasso.cv$lambda.1se)$beta as.matrix(cv_betas)[as.matrix(cv_betas)>0,] ``` ``` ## bmi map ltg bmi^2 glu^2 age:sex ## 502.887614 205.892655 453.531965 17.266008 39.776736 60.581213 ## age:map age:glu bmi:map ## 14.945884 7.108415 59.023875 ``` --- class: eg ### Diabetes example We can choose between the two competing models using **cross validation**. ```r library(caret) control <- trainControl(method = "cv", number = 10) dat <- cbind(y, x) aic_cv <- train(y ~ bmi + map + ltg + glu + `age^2` + `bmi^2` + `glu^2` + `age:sex` + `age:map` + `age:ltg` + `age:glu` + `sex:map` + `bmi:map`, method = "lm", data = dat, trControl = control) cv_cv <- train(y ~ bmi + map + ltg + `bmi^2` + `glu^2` + `age:sex` + `age:map` + `age:glu` + `bmi:map`, method = "lm", data = dat, trControl = control) results <- resamples(list(AIC = aic_cv, CV = cv_cv)) ggplot(results) + theme_classic(base_size = 18) + labs(x = "MAE") ``` <img src="lecB_files/figure-html/unnamed-chunk-6-1.png" width="864" /> --- class: segue # B2. The mplot package --- ## Installing the mplot package ```r # install.packages("devtools") # devtools::install_github("garthtarr/mplot") install.packages("mplot") library(mplot) ``` Website: http://garthtarr.github.io/mplot ### Main functions * `af()` for the adaptive fence for `lm()` and `glm()` * `vis()` for variable importance plots and model stability plots for `lm()` and `glm()` * `bglmnet()` for bootstrapping `glmnet()` and variable importance and model stability plots * `mplot()` for an interactive shiny interface .footnote[ Tarr, Müller, and Welsh (2018) ] --- ## Motivation – Recall Anscombe’s quartet <img src="lecB_files/figure-html/anscombe-1.png" style="display: block; margin: auto;" /> --- ## Datasaurus ```r library(datasauRus) ``` <!-- --> .footnote[ Matejka J and Fitzmaurice G (2017). [Same Stats, Different Graphs: Generating Datasets with Varied Appearance and Identical Statistics through Simulated Annealing.](https://www.autodeskresearch.com/publications/samestats) Autodesk Research, Toronto Ontario Canada. DOI: 10.1145/3025453.3025912 ] --- ## See also <!-- --> 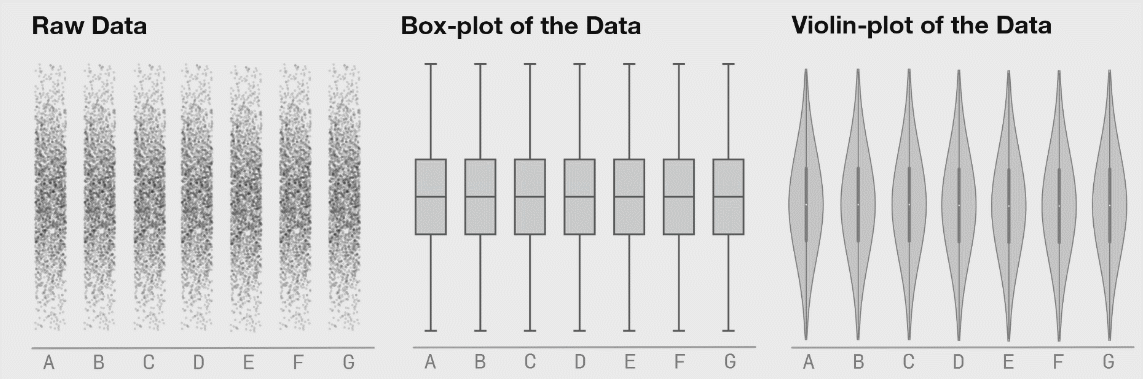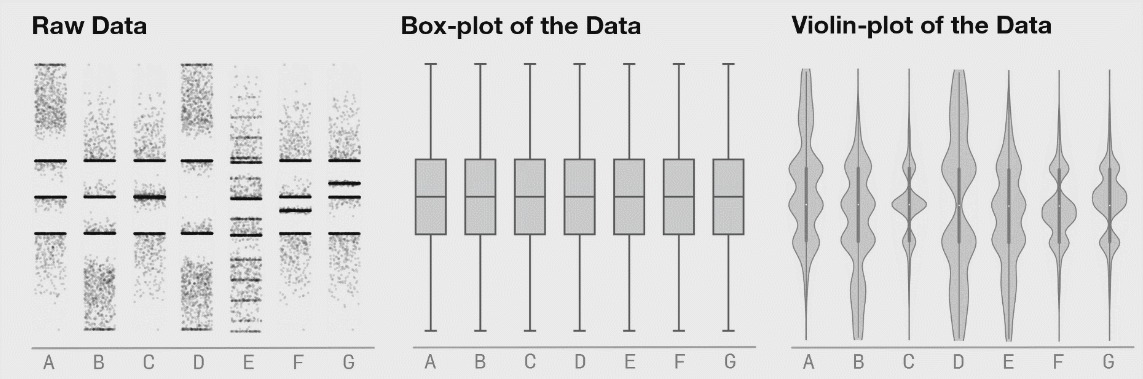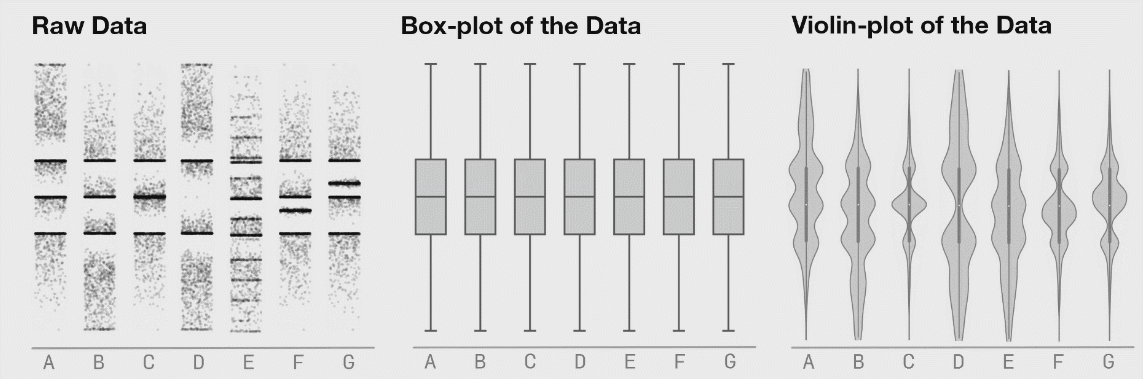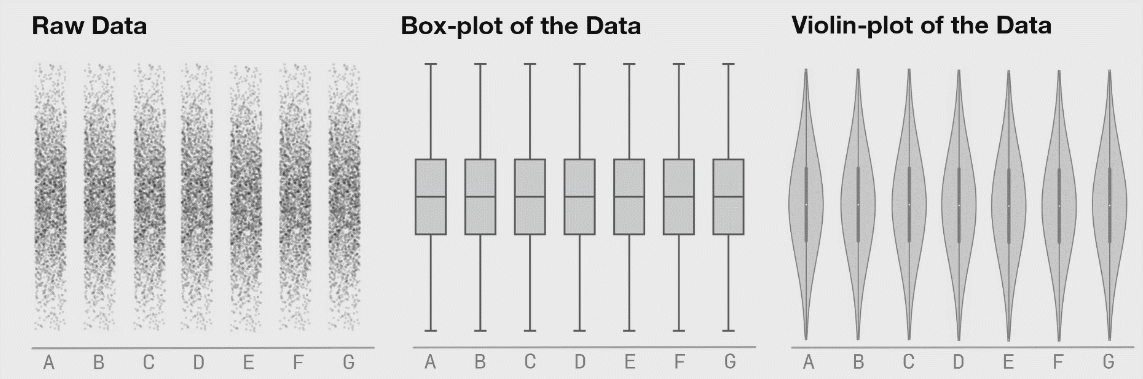<!-- --> .footnote[ Matejka J and Fitzmaurice G (2017). [Same Stats, Different Graphs: Generating Datasets with Varied Appearance and Identical Statistics through Simulated Annealing.](https://www.autodeskresearch.com/publications/samestats) Autodesk Research, Toronto Ontario Canada. DOI: 10.1145/3025453.3025912 ] --- ## Graphical tools for model selection * .red[Underused!] * .blue[Fence] and its variants (series of papers by Jiang et al, starting with Jiang, Rao, Gu, and Nguyen (2008)) * .blue[Stability selection] Meinshausen and Bühlmann (2010) * .blue[Model selection curves] Müller and Welsh (2010) * .blue[Variable inclusion plots] Müller and Welsh (2010) and Murray, Heritier, and Müller (2013) * .red[Variable selection with `mplot`] (this lecture) --- class: segue # Artificial example --- class: eg ## Artificial example: Correlations <img src="lecB_files/figure-html/artificialcorrelations-1.png" style="display: block; margin: auto;" /> --- class: eg ## Artificial example: Full model .scroll-output[ ```r library(mplot) lm.art = lm(y ~ ., data = artificialeg) summary(lm.art) ``` ``` ## ## Call: ## lm(formula = y ~ ., data = artificialeg) ## ## Residuals: ## Min 1Q Median 3Q Max ## -4.7254 -1.2126 0.2456 1.3446 3.4808 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) -0.1002 0.3268 -0.307 0.761 ## x1 0.6403 0.6924 0.925 0.361 ## x2 0.2574 0.6176 0.417 0.679 ## x3 -0.5105 1.2391 -0.412 0.683 ## x4 -0.2978 0.2515 -1.184 0.243 ## x5 0.3551 0.5982 0.594 0.556 ## x6 -0.5428 0.9612 -0.565 0.575 ## x7 -0.4280 0.6308 -0.678 0.501 ## x8 0.1503 0.6175 0.243 0.809 ## x9 0.3997 0.6379 0.627 0.535 ## ## Residual standard error: 2.023 on 40 degrees of freedom ## Multiple R-squared: 0.7508, Adjusted R-squared: 0.6947 ## F-statistic: 13.39 on 9 and 40 DF, p-value: 1.476e-09 ``` ] --- class: eg ## Artificial example: Final model .scroll-output[ ```r step(lm.art) ``` ``` ## Start: AIC=79.3 ## y ~ x1 + x2 + x3 + x4 + x5 + x6 + x7 + x8 + x9 ## ## Df Sum of Sq RSS AIC ## - x8 1 0.2423 163.94 77.374 ## - x3 1 0.6946 164.39 77.512 ## - x2 1 0.7107 164.41 77.517 ## - x6 1 1.3051 165.00 77.698 ## - x5 1 1.4425 165.14 77.739 ## - x9 1 1.6065 165.31 77.789 ## - x7 1 1.8835 165.58 77.873 ## - x1 1 3.4999 167.20 78.358 ## - x4 1 5.7367 169.44 79.023 ## <none> 163.70 79.301 ## ## Step: AIC=77.37 ## y ~ x1 + x2 + x3 + x4 + x5 + x6 + x7 + x9 ## ## Df Sum of Sq RSS AIC ## <none> 163.94 77.374 ## - x2 1 20.359 184.30 81.227 ## - x5 1 25.966 189.91 82.726 ## - x9 1 33.607 197.55 84.698 ## - x4 1 34.504 198.45 84.925 ## - x7 1 62.097 226.04 91.434 ## - x1 1 68.253 232.19 92.778 ## - x3 1 71.301 235.24 93.430 ## - x6 1 107.873 271.81 100.655 ``` ``` ## ## Call: ## lm(formula = y ~ x1 + x2 + x3 + x4 + x5 + x6 + x7 + x9, data = artificialeg) ## ## Coefficients: ## (Intercept) x1 x2 x3 x4 ## -0.1143 0.8019 0.4011 -0.8083 -0.3514 ## x5 x6 x7 x9 ## 0.4927 -0.7738 -0.5772 0.5478 ``` ] --- class: eg ## Artificial example: Final model .scroll-output[ ```r lm.fin = lm(y ~ . - x8, data = artificialeg) summary(lm.fin) ``` ``` ## ## Call: ## lm(formula = y ~ . - x8, data = artificialeg) ## ## Residuals: ## Min 1Q Median 3Q Max ## -4.6881 -1.2352 0.2787 1.4102 3.5420 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) -0.1143 0.3179 -0.360 0.720952 ## x1 0.8019 0.1941 4.132 0.000173 *** ## x2 0.4011 0.1778 2.256 0.029438 * ## x3 -0.8083 0.1914 -4.223 0.000131 *** ## x4 -0.3514 0.1196 -2.938 0.005408 ** ## x5 0.4927 0.1933 2.548 0.014667 * ## x6 -0.7738 0.1490 -5.194 6e-06 *** ## x7 -0.5772 0.1465 -3.941 0.000309 *** ## x9 0.5478 0.1890 2.899 0.005987 ** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 2 on 41 degrees of freedom ## Multiple R-squared: 0.7504, Adjusted R-squared: 0.7017 ## F-statistic: 15.41 on 8 and 41 DF, p-value: 3.692e-10 ``` ] --- class: eg ## Artificial example: Data generating model The .red[true data generating model] is: `\(\color{red}{y} = 0.6\,\color{red}{x_8} + \color{red}{\varepsilon}\)`. ```r art.true = lm(y ~ x8, data = artificialeg) summary(art.true) ``` ``` ## ## Call: ## lm(formula = y ~ x8, data = artificialeg) ## ## Residuals: ## Min 1Q Median 3Q Max ## -5.6013 -1.2985 0.1303 1.4605 3.8056 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 0.03235 0.29458 0.11 0.913 ## x8 0.54928 0.05264 10.43 6.18e-14 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 2.046 on 48 degrees of freedom ## Multiple R-squared: 0.694, Adjusted R-squared: 0.6877 ## F-statistic: 108.9 on 1 and 48 DF, p-value: 6.18e-14 ``` --- class: segue # Information criteria can be blind .blockquote[ Use resampling and graphs to **see** more! ] --- ## Bootstrapping regression models .blockquote[ The **bootstrap** is a resampling strategy to generate `\(B\)` training sets ] <br> ### Popular implementations - **Paired bootstrap**: samples (with replacement) rows of `\([\boldsymbol{y},\boldsymbol{X}]\)` - .blue[**Weighted bootstrap**] (used in `vis()` and `bglmnet()`): mimics the paired bootstrap by keeping `\([\boldsymbol{y},\boldsymbol{X}]\)` as is but weighs each observation (row) through using a weights vector, where the weights are sampled from a distribution with expectation 1 (e.g. exponential). - .blue[**Parametric bootstrap**] (used in `af()`): estimate the (parametric) distribution of `\(\boldsymbol{y}\)` (or `\(\boldsymbol{\varepsilon}\)`) and draw new observations from it - **Residual bootstrap**: baseline model + bootstrapped error --- ## Stability - In practice it is important that selected models are stable .blockquote[ **Key idea**: small changes should have small effects ] - Example: Information criterion is *unstable* when `\(\hat{\alpha}(\lambda)\)` is selected with dimension `\(p_{\hat{\alpha}(\lambda)}\)` but, for some small `\(\delta>0\)`, `\(\hat{\alpha}(\lambda+\delta)\)` selected with `\(p_{\hat{\alpha}(\lambda+\delta)} < p_{\hat{\alpha}(\lambda)}\)` - Investigate model selection criterion in a neighborhood of `\(\lambda\)` --- ## Obtaining stability information .blockquote[ **Use resampling**: repeatedly calculate how well (the best) model(s) perform given `\(\lambda\)`, summarise performance with statistics and graphs ] <br> ### We will do the following: * Investigate model detectability as a function of `\(\lambda\)` * Estimate probability that model `\(\alpha\in\mathcal{A}\)` is optimal at `\(\lambda\)` * Estimate best model(s) within each size/dimension * Show model selection curves also for models that are never optimal --- class: segue # Variable inclusion plots --- ## Variable inclusion plots .blockquote[ **Aim**: To visualise inclusion probabilities as a function of the penalty multiplier `\(\lambda \in [0,2\log(n)]\)` ] ### Procedure 1. Calculate (weighted) .red[bootstrap samples] `\(\color{red}{b}=1,\ldots,B\)`. 2. For each .red[bootstrap sample], at each `\(\color{blue}{\lambda}\)` .blue[value], find `\(\hat{\alpha}_{\color{blue}{\lambda}}^{(\color{red}{b})}\in\mathcal{A}\)` as the model with smallest `\(\text{GIC}(\alpha;\color{blue}{\lambda}) = -2\times \text{LogLik}(\alpha)+\color{blue}{\lambda} p_\alpha\)`. 3. The **inclusion probability** for variable `\(x_j\)` is estimated as `\(\frac{1}{B}\sum_{\color{red}{b}=1}^B 1\{j\in \hat{\alpha}_{\color{blue}{\lambda}}^{(\color{red}{b})}\}\)`. .blockquote[ The **inclusion probability** for variable `\(x_j\)` is the proportion of times variable `\(x_j\)` is in the best model ] .footnote[ - Müller and Welsh (2010) for linear regression models - Murray, Heritier, and Müller (2013) for generalised linear models ] --- class: eg ## Artificial example: VIP ```r library(mplot) vis.art = vis(lm.art) plot(vis.art, which = "vip", interactive = TRUE) ``` <iframe src = "src/artvisplot1.html" width = "100%" height = "90%" frameborder="0"></iframe> --- class: segue # Model stability plots --- class: eg ## Artificial example: Loss against size ```r plot(vis.art, which="lvk", interactive = TRUE) ``` <iframe src = "src/artvisredplot1.html" width = "100%" height = "90%" frameborder="0"></iframe> --- class: eg ## Artificial example: Loss against size ```r plot(vis.art, which = "lvk", highlight = "x6", interactive = TRUE) ``` <iframe src = "src/artvisredplot2.html" width = "100%" height = "90%" frameborder="0"></iframe> --- ## Model stability plots .blockquote[ **Aim**: To add value to the loss against size plots by choosing a symbol size proportional to a measure of stability ] <br> ### Procedure 1. Calculate (weighted) bootstrap samples `\(b=1,\ldots,B\)` 2. For each bootstrap sample, identify the *best* model at each dimension 3. Add this information to the loss against size plot using model identifiers that are proportional to the frequency with which a model was identified as being *best* at each model size .footnote[ Murray, Heritier, and Müller (2013) for generalised linear models ] --- class: eg ## Artificial example: Model stability plot ```r plot(vis.art, which = "boot", interactive = FALSE) ``` <!-- --> --- ## Artificial example: Model stability plot ```r plot(vis.art, which = "boot", interactive = TRUE) ``` <iframe src = "src/artmspred2.html" width = "100%" height = "90%" frameborder="0"></iframe> --- class: segue # Bootstrapping the lasso --- class: eg ## Artificial example: Lasso ```r library(lars) x = as.matrix(subset(artificialeg, select = -y)) y = as.matrix(subset(artificialeg, select = y)) art.lars=lars(x, y) plot(art.lars, xvar = "step", breaks = FALSE, lwd = 2, cex.lab = 1.4) ``` <!-- --> --- class: eg ## Artificial example: Lasso ```r plot(art.lars, xvar = "step", plottype = "Cp", lwd = 2, cex.lab = 1.4) ``` <!-- --> --- class: eg ## Artificial example: Lasso <!-- --> --- class: eg ## Artificial example: Lasso (zoomed) <!-- --> --- class: eg ## Bootstrapping the lasso ```r bgn.art = bglmnet(lm.art, lambda = seq(0.05, 2.75, 0.1)) plot(bgn.art, which = "boot", interactive = TRUE) ``` <iframe src = "src/bglmnetplot1.html" width = "100%" height = "90%" frameborder="0"></iframe> --- class: eg ## Bootstrapping the lasso ```r bgn.art = bglmnet(lm.art) plot(bgn.art, which = "vip", interactive = TRUE) ``` <iframe src = "src/bglmnetplot2.html" width = "100%" height = "90%" frameborder="0"></iframe> --- class: segue # B3. Subtractive stability measures # `\(\hat{Q}(M) = s - \sum_{j\in M}s_j\)` --- ## Stability measures - Use .red[resampling], say `\(B\)` resamples (100, 1000 or more) - Consider .blue[baseline models] (e.g. all models on a solution path), say `\(K\)` such models - Learn from an array of `\(K\times p\times B\)` regression coefficients - Calculate for each feature `\(j=1,\ldots,p\)` - Inclusion frequency - Exclusion frequency - **Other variable importance measure** --- ## What is a subtractive measure? Ingenious idea by Jiang, Nguyen and Rao (2011, Statistics and Its Interface) A **subtractive measure** `\(\hat{Q}\)` satisfies `$$\hat{Q}(M_K) = s - \sum_{j\in M_K}s_j$$` with `\(s_j \geq 0\)`, `\(k = 1,\ldots, K\)`. Think .red[.bold["lack-of-fit"]] measure (like a log-likelihood) with .red[.bold["benefits"]] (something additional to a log-likelihood). --- class: eg ## Toy example - Let the full model be `\(\{1,2,3,4,5,6,7,8,9,10\}\)`, i.e. `\(p=10\)` - Let's assume that `\(s_1 = |-0.05|\)`, `\(s_2 =|-0.19|\)`, with remaining `\(s_j\)`'s being 0.34, 0.21, 0.30, 0.19, 0.02, 0.09, **0.42** and 0.03 - Note: all `\(s_j\)` values are non-negative - Let `\(s = \sum s_j = 1.84\)` - That model minimizing `\(\hat{Q}\)` over all models with one variable is `\(\{9\}\)` - The best model with two variables is `\(\{3,9\}\)` - `\(\hat{Q} \geq 0\)` by construction, with equality for the full model --- ## What are the benefits? Magic in `\(s_j\)` and constructing `\(\hat{Q}\)` - Fast access to identify best model of size `\(d\)` - Remains fast when bootstrapping Full model fit sufficient to get well performing `\(s_j\)`'s - `\(s_j = |\hat{\beta}_j|\)` (.red[robust], .red[penalised]) - `\(s_j = |t_j|\)` (standardised coefficients) - .red[Robust quasi-deviance statistics] (differences to full/large baseline model) --- ## Coming soon to the mplot package - Resample `\(\{y,\mathbf{X}\}\)`, e.g. through weighted bootstrap - Use OLS (LIMMA), Ridge, Lasso, Elastic-net, etc - Observe `\(s_j^{(b)}\)`, `\(b = 1,\ldots,B\)`, `\(j = 1,\ldots, p\)` - Find the empirical probability that `\(s_j>s_k\)` - Derive new indices - Mean bootstrapped rank of `\(s_j^{(b)}\)` - Standard deviation of `\(\text{rank}(s_j^{(b)})\)` - Devise new visualisations - Preprints: Smith, Günnewig, and Müller (2020) and Smith, Müller, and Günnewig (2020) --- ## A function to bootstrap the full model ```r sstab = function(mf, B = 100) { full_coeff = coefficients(mf) kf = length(full_coeff) coef.res = matrix(ncol = kf, nrow = B) colnames(coef.res) = names(full_coeff) formula = stats::formula(mf) data = model.frame(mf) for(i in 1:B){ wts = stats::rexp(n = length(resid(mf)), rate = 1) data$wts = wts mod = stats::lm(formula = formula, data = data, weights = wts) coef.res[i, ] = coefficients(mod) } return(coef.res) } ``` --- class: eg ## Diabetes example ```r data("diabetes", package = "lars") x = diabetes$x y = diabetes$y df = data.frame(scale(cbind(y, x))) lm1 = lm(y ~ ., data = df) sj = sstab(lm1) round(head(sj[, -1], n = 10), 2) ``` ``` ## age sex bmi map tc ldl hdl tch ltg glu ## [1,] -0.05 -0.19 0.34 0.21 -0.30 0.19 -0.02 0.09 0.42 0.03 ## [2,] 0.02 -0.16 0.29 0.23 -0.21 -0.04 0.08 0.29 0.29 0.02 ## [3,] 0.02 -0.04 0.26 0.17 -0.37 0.20 0.05 0.20 0.43 0.06 ## [4,] 0.00 -0.15 0.32 0.26 -0.39 0.11 0.04 0.16 0.40 0.01 ## [5,] -0.04 -0.17 0.34 0.21 -0.56 0.30 0.10 0.15 0.50 0.06 ## [6,] -0.05 -0.11 0.29 0.18 -0.64 0.47 0.11 0.06 0.50 0.08 ## [7,] 0.01 -0.17 0.28 0.23 -0.31 0.03 -0.05 0.14 0.41 0.08 ## [8,] 0.00 -0.12 0.37 0.15 -0.24 0.06 -0.09 0.06 0.34 0.07 ## [9,] 0.00 -0.04 0.33 0.14 -0.40 0.20 0.11 0.08 0.50 0.08 ## [10,] 0.01 -0.17 0.34 0.24 -0.76 0.48 0.17 0.15 0.56 -0.01 ``` --- class: eg ## Diabetes example ```r sj = abs(sj[, -1]) sj_ranks = apply(sj, 1, rank) head(t(sj_ranks), n = 10) ``` ``` ## age sex bmi map tc ldl hdl tch ltg glu ## [1,] 3 6 9 7 8 5 1 4 10 2 ## [2,] 2 5 9 7 6 3 4 8 10 1 ## [3,] 1 2 8 5 9 7 3 6 10 4 ## [4,] 1 5 8 7 9 4 3 6 10 2 ## [5,] 1 5 8 6 10 7 3 4 9 2 ## [6,] 1 5 7 6 10 8 4 2 9 3 ## [7,] 1 6 8 7 9 2 3 5 10 4 ## [8,] 1 6 10 7 8 2 5 3 9 4 ## [9,] 1 2 8 6 9 7 5 3 10 4 ## [10,] 1 4 7 6 10 8 5 3 9 2 ``` --- class: eg ## Diabetes example ```r sj_rank_mean = sort(apply(sj_ranks, 1, mean), decreasing = TRUE) sj_rank_mean ``` ``` ## ltg tc bmi map ldl sex tch hdl glu age ## 9.33 8.81 8.06 6.21 6.21 4.67 4.33 3.40 2.38 1.60 ``` ```r # tc table(sj_ranks[5, ])/100 ``` ``` ## ## 1 3 4 5 6 7 8 9 10 ## 0.02 0.01 0.02 0.02 0.02 0.02 0.18 0.22 0.49 ``` ```r # ltg table(sj_ranks[9, ])/100 ``` ``` ## ## 6 8 9 10 ## 0.01 0.03 0.57 0.39 ``` --- class: font100 columns-2 ## References <a name=bib-Jiang2008></a>[Jiang, J, J. S. Rao, Z. Gu, et al.](#cite-Jiang2008) (2008). "Fence methods for mixed model selection". In: _The Annals of Statistics_ 36.4, pp. 1669-1692. DOI: [10.1214/07-AOS517](https://doi.org/10.1214%2F07-AOS517). <a name=bib-Meinshausen2010></a>[Meinshausen, N. and P. Bühlmann](#cite-Meinshausen2010) (2010). "Stability selection". In: _Journal of the Royal Statistical Society. Series B. Statistical Methodology_ 72.4, pp. 417-473. DOI: [10.1111/j.1467-9868.2010.00740.x](https://doi.org/10.1111%2Fj.1467-9868.2010.00740.x). <a name=bib-Mueller2010></a>[Müller, S. and A. H. Welsh](#cite-Mueller2010) (2010). "On Model Selection Curves". In: _International Statistical Review_ 78.2, pp. 240-256. DOI: [10.1111/j.1751-5823.2010.00108.x](https://doi.org/10.1111%2Fj.1751-5823.2010.00108.x). <a name=bib-Murray2013></a>[Murray, K, S. Heritier, and S. Müller](#cite-Murray2013) (2013). "Graphical tools for model selection in generalized linear models". In: _Statistics in Medicine_ 32.25, pp. 4438-4451. DOI: [10.1002/sim.5855](https://doi.org/10.1002%2Fsim.5855). <a name=bib-Smith2020a></a>[Smith, C, B. Günnewig, and S. Müller](#cite-Smith2020a) (2020). "Robust subtractive stability measures for fast and exhaustive feature importance ranking and selection in generalized linear models". In: _Preprint_, pp. 1-27. <a name=bib-Smith2020b></a>[Smith, C, S. Müller, and B. Günnewig](#cite-Smith2020b) (2020). "Visualization of variable importance differences". In: _Preprint_, pp. 1-8. <a name=bib-Tarr2018></a>[Tarr, G, S. Müller, and A. H. Welsh](#cite-Tarr2018) (2018). "mplot: An R Package for Graphical Model Stability and Variable Selection Procedures". In: _Journal of Statistical Software_ 83.9, pp. 1-28. DOI: [10.18637/jss.v083.i09](https://doi.org/10.18637%2Fjss.v083.i09). --- class: code50 columns-2 ```r devtools::session_info(include_base = FALSE) ``` ``` ## ─ Session info ─────────────────────────────────────────────────────────────────────────────────── ## setting value ## version R version 4.0.0 (2020-04-24) ## os macOS Catalina 10.15.5 ## system x86_64, darwin17.0 ## ui X11 ## language (EN) ## collate en_AU.UTF-8 ## ctype en_AU.UTF-8 ## tz Australia/Sydney ## date 2020-07-16 ## ## ─ Packages ─────────────────────────────────────────────────────────────────────────────────────── ## package * version date lib source ## assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.0.0) ## backports 1.1.8 2020-06-17 [1] CRAN (R 4.0.0) ## bibtex 0.4.2.2 2020-01-02 [1] CRAN (R 4.0.0) ## callr 3.4.3 2020-03-28 [1] CRAN (R 4.0.0) ## caret * 6.0-86 2020-03-20 [1] CRAN (R 4.0.0) ## class 7.3-17 2020-04-26 [1] CRAN (R 4.0.0) ## cli 2.0.2 2020-02-28 [1] CRAN (R 4.0.0) ## codetools 0.2-16 2018-12-24 [1] CRAN (R 4.0.0) ## colorspace 1.4-1 2019-03-18 [1] CRAN (R 4.0.0) ## crayon 1.3.4 2017-09-16 [1] CRAN (R 4.0.0) ## data.table 1.12.8 2019-12-09 [1] CRAN (R 4.0.0) ## datasauRus * 0.1.4 2018-09-20 [1] CRAN (R 4.0.0) ## desc 1.2.0 2018-05-01 [1] CRAN (R 4.0.0) ## devtools 2.3.0 2020-04-10 [1] CRAN (R 4.0.0) ## digest 0.6.25 2020-02-23 [1] CRAN (R 4.0.0) ## doParallel 1.0.15 2019-08-02 [1] CRAN (R 4.0.0) ## doRNG 1.8.2 2020-01-27 [1] CRAN (R 4.0.0) ## dplyr 1.0.0 2020-05-29 [1] CRAN (R 4.0.0) ## ellipsis 0.3.1 2020-05-15 [1] CRAN (R 4.0.0) ## evaluate 0.14 2019-05-28 [1] CRAN (R 4.0.0) ## fansi 0.4.1 2020-01-08 [1] CRAN (R 4.0.0) ## farver 2.0.3 2020-01-16 [1] CRAN (R 4.0.0) ## fastmap 1.0.1 2019-10-08 [1] CRAN (R 4.0.0) ## foreach 1.5.0 2020-03-30 [1] CRAN (R 4.0.0) ## fs 1.4.1 2020-04-04 [1] CRAN (R 4.0.0) ## generics 0.0.2 2018-11-29 [1] CRAN (R 4.0.0) ## ggplot2 * 3.3.2 2020-06-19 [1] CRAN (R 4.0.0) ## glmnet * 4.0-2 2020-06-16 [1] CRAN (R 4.0.0) ## glue 1.4.1 2020-05-13 [1] CRAN (R 4.0.0) ## googleVis 0.6.5 2020-06-08 [1] CRAN (R 4.0.0) ## gower 0.2.2 2020-06-23 [1] CRAN (R 4.0.0) ## gtable 0.3.0 2019-03-25 [1] CRAN (R 4.0.0) ## htmltools 0.5.0 2020-06-16 [1] CRAN (R 4.0.0) ## httpuv 1.5.4 2020-06-06 [1] CRAN (R 4.0.0) ## httr 1.4.1 2019-08-05 [1] CRAN (R 4.0.0) ## ipred 0.9-9 2019-04-28 [1] CRAN (R 4.0.0) ## iterators 1.0.12 2019-07-26 [1] CRAN (R 4.0.0) ## jsonlite 1.7.0 2020-06-25 [1] CRAN (R 4.0.0) ## knitcitations * 1.0.10 2019-09-15 [1] CRAN (R 4.0.0) ## knitr 1.29 2020-06-23 [1] CRAN (R 4.0.0) ## labeling 0.3 2014-08-23 [1] CRAN (R 4.0.0) ## lars * 1.2 2013-04-24 [1] CRAN (R 4.0.0) ## later 1.1.0.1 2020-06-05 [1] CRAN (R 4.0.0) ## lattice * 0.20-41 2020-04-02 [1] CRAN (R 4.0.0) ## lava 1.6.7 2020-03-05 [1] CRAN (R 4.0.0) ## leaps 3.1 2020-01-16 [1] CRAN (R 4.0.0) ## lifecycle 0.2.0 2020-03-06 [1] CRAN (R 4.0.0) ## lubridate 1.7.9 2020-06-08 [1] CRAN (R 4.0.0) ## magrittr 1.5 2014-11-22 [1] CRAN (R 4.0.0) ## MASS 7.3-51.6 2020-04-26 [1] CRAN (R 4.0.0) ## Matrix * 1.2-18 2019-11-27 [1] CRAN (R 4.0.0) ## memoise 1.1.0 2017-04-21 [1] CRAN (R 4.0.0) ## mime 0.9 2020-02-04 [1] CRAN (R 4.0.0) ## ModelMetrics 1.2.2.2 2020-03-17 [1] CRAN (R 4.0.0) ## mplot * 1.0.4 2020-02-15 [1] CRAN (R 4.0.0) ## munsell 0.5.0 2018-06-12 [1] CRAN (R 4.0.0) ## nlme 3.1-148 2020-05-24 [1] CRAN (R 4.0.0) ## nnet 7.3-14 2020-04-26 [1] CRAN (R 4.0.0) ## pillar 1.4.4 2020-05-05 [1] CRAN (R 4.0.0) ## pkgbuild 1.0.8 2020-05-07 [1] CRAN (R 4.0.0) ## pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.0.0) ## pkgload 1.1.0 2020-05-29 [1] CRAN (R 4.0.0) ## plyr 1.8.6 2020-03-03 [1] CRAN (R 4.0.0) ## prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.0.0) ## pROC 1.16.2 2020-03-19 [1] CRAN (R 4.0.0) ## processx 3.4.2 2020-02-09 [1] CRAN (R 4.0.0) ## prodlim 2019.11.13 2019-11-17 [1] CRAN (R 4.0.0) ## promises 1.1.1 2020-06-09 [1] CRAN (R 4.0.0) ## ps 1.3.3 2020-05-08 [1] CRAN (R 4.0.0) ## purrr 0.3.4 2020-04-17 [1] CRAN (R 4.0.0) ## R6 2.4.1 2019-11-12 [1] CRAN (R 4.0.0) ## Rcpp 1.0.4.6 2020-04-09 [1] CRAN (R 4.0.0) ## recipes 0.1.13 2020-06-23 [1] CRAN (R 4.0.0) ## RefManageR 1.2.12 2019-04-03 [1] CRAN (R 4.0.0) ## remotes 2.1.1 2020-02-15 [1] CRAN (R 4.0.0) ## reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.0.0) ## rlang 0.4.6 2020-05-02 [1] CRAN (R 4.0.0) ## rmarkdown 2.3 2020-06-18 [1] CRAN (R 4.0.0) ## rngtools 1.5 2020-01-23 [1] CRAN (R 4.0.0) ## rpart 4.1-15 2019-04-12 [1] CRAN (R 4.0.0) ## rprojroot 1.3-2 2018-01-03 [1] CRAN (R 4.0.0) ## scales 1.1.1 2020-05-11 [1] CRAN (R 4.0.0) ## sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.0.0) ## shape 1.4.4 2018-02-07 [1] CRAN (R 4.0.0) ## shiny 1.5.0 2020-06-23 [1] CRAN (R 4.0.0) ## shinydashboard 0.7.1 2018-10-17 [1] CRAN (R 4.0.0) ## stringi 1.4.6 2020-02-17 [1] CRAN (R 4.0.0) ## stringr 1.4.0 2019-02-10 [1] CRAN (R 4.0.0) ## survival 3.2-3 2020-06-13 [1] CRAN (R 4.0.0) ## testthat 2.3.2 2020-03-02 [1] CRAN (R 4.0.0) ## tibble 3.0.1 2020-04-20 [1] CRAN (R 4.0.0) ## tidyselect 1.1.0 2020-05-11 [1] CRAN (R 4.0.0) ## timeDate 3043.102 2018-02-21 [1] CRAN (R 4.0.0) ## usethis 1.6.1 2020-04-29 [1] CRAN (R 4.0.0) ## vctrs 0.3.1 2020-06-05 [1] CRAN (R 4.0.0) ## withr 2.2.0 2020-04-20 [1] CRAN (R 4.0.0) ## xaringan 0.16 2020-03-31 [1] CRAN (R 4.0.0) ## xfun 0.15 2020-06-21 [1] CRAN (R 4.0.0) ## xml2 1.3.2 2020-04-23 [1] CRAN (R 4.0.0) ## xtable 1.8-4 2019-04-21 [1] CRAN (R 4.0.0) ## yaml 2.2.1 2020-02-01 [1] CRAN (R 4.0.0) ## ## [1] /Library/Frameworks/R.framework/Versions/4.0/Resources/library ```