Lab 2: Exhaustive searching and GLMs
Samuel Müller and Garth Tarr
Learning outcomes
- Marginality principle
- Exhaustive search for linear models
- Exhaustive search for generalised linear models
Required R packages:
install.packages(c("mplot", "leaps", "bestglm", "car"))Crime data
data("UScrime", package = "MASS")
M0 = lm(y ~ 1, data = UScrime) # Null model
M1 = lm(y ~ ., data = UScrime) # Full modelExercises
- Use the
regsubsets()function from theleapspackage to obtain the 50 best models for each model size and plot their RSS against model dimension.
library(leaps)
rgsbst.out = regsubsets(y ~ ., data = UScrime, nbest = 50, nvmax = NULL,
method = "exhaustive", really.big = TRUE)
rss = summary(rgsbst.out)$rss
p = rowSums(summary(rgsbst.out)$which)
plot(rss ~ p)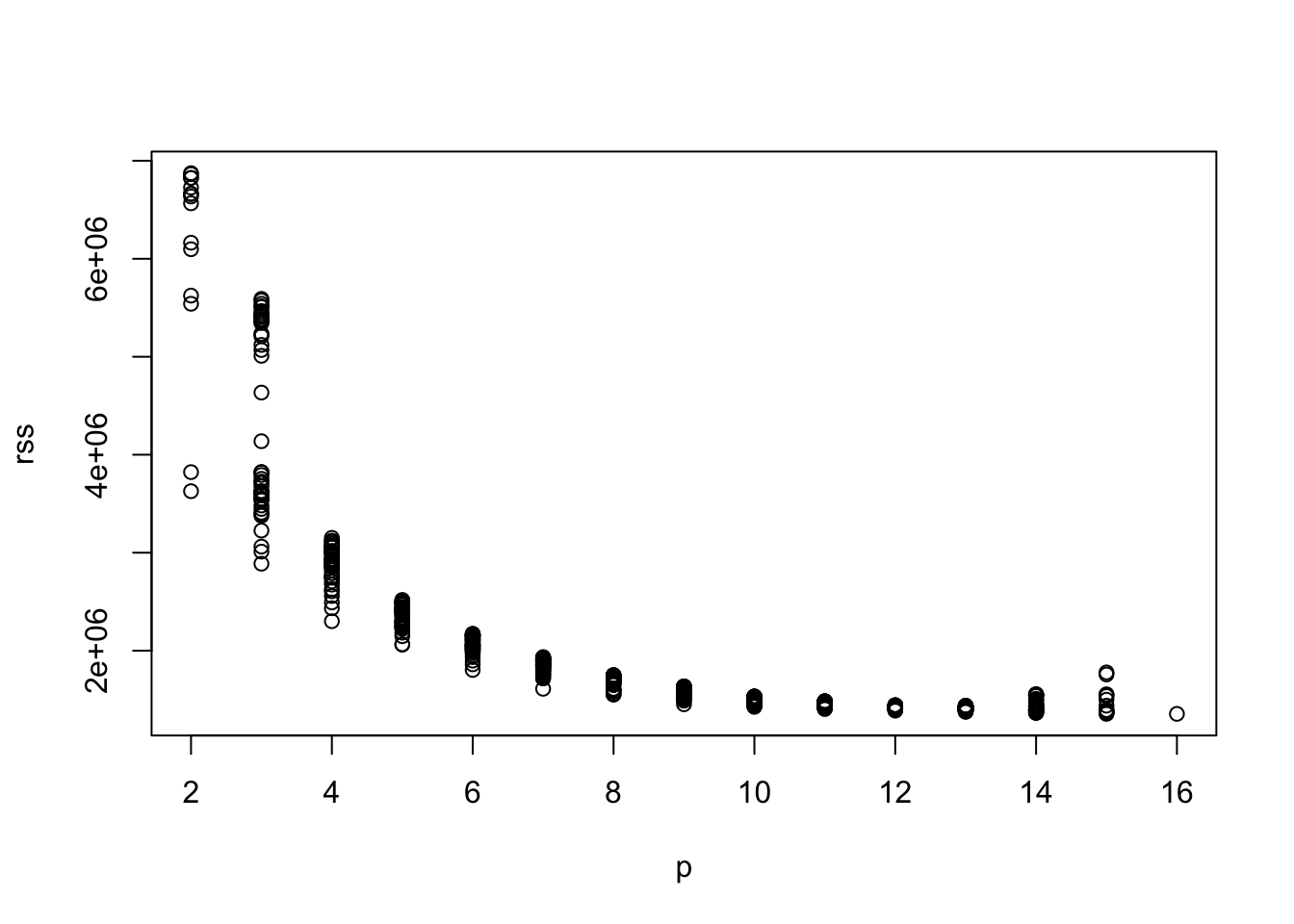
- Use the
method = "forward"andmethod = "backward"andmethod = "exhaustive"options to perform forward, backward and exhaustive model selection and compare the results (only find a single best model at each dimension). Hint: Theregsubsetsfunction returns several information criteria, choose for example Mallow’s Cp.
fwd = regsubsets(y ~ ., data = UScrime, nvmax = NULL, method = "forward")
bwd = regsubsets(y ~ ., data = UScrime, nvmax = NULL, method = "backward")
exh = regsubsets(y ~ ., data = UScrime, nvmax = NULL, method = "exhaustive")
as.data.frame(summary(fwd)$outmat)## M So Ed Po1 Po2 LF M.F Pop NW U1 U2 GDP Ineq Prob
## 1 ( 1 ) *
## 2 ( 1 ) * *
## 3 ( 1 ) * * *
## 4 ( 1 ) * * * *
## 5 ( 1 ) * * * * *
## 6 ( 1 ) * * * * * *
## 7 ( 1 ) * * * * * * *
## 8 ( 1 ) * * * * * * * *
## 9 ( 1 ) * * * * * * * * *
## 10 ( 1 ) * * * * * * * * * *
## 11 ( 1 ) * * * * * * * * * * *
## 12 ( 1 ) * * * * * * * * * * * *
## 13 ( 1 ) * * * * * * * * * * * * *
## 14 ( 1 ) * * * * * * * * * * * * *
## 15 ( 1 ) * * * * * * * * * * * * * *
## Time
## 1 ( 1 )
## 2 ( 1 )
## 3 ( 1 )
## 4 ( 1 )
## 5 ( 1 )
## 6 ( 1 )
## 7 ( 1 )
## 8 ( 1 )
## 9 ( 1 )
## 10 ( 1 )
## 11 ( 1 )
## 12 ( 1 )
## 13 ( 1 )
## 14 ( 1 ) *
## 15 ( 1 ) *as.data.frame(summary(bwd)$outmat)## M So Ed Po1 Po2 LF M.F Pop NW U1 U2 GDP Ineq Prob
## 1 ( 1 ) *
## 2 ( 1 ) * *
## 3 ( 1 ) * * *
## 4 ( 1 ) * * * *
## 5 ( 1 ) * * * * *
## 6 ( 1 ) * * * * * *
## 7 ( 1 ) * * * * * * *
## 8 ( 1 ) * * * * * * * *
## 9 ( 1 ) * * * * * * * * *
## 10 ( 1 ) * * * * * * * * * *
## 11 ( 1 ) * * * * * * * * * * *
## 12 ( 1 ) * * * * * * * * * * * *
## 13 ( 1 ) * * * * * * * * * * * * *
## 14 ( 1 ) * * * * * * * * * * * * *
## 15 ( 1 ) * * * * * * * * * * * * * *
## Time
## 1 ( 1 )
## 2 ( 1 )
## 3 ( 1 )
## 4 ( 1 )
## 5 ( 1 )
## 6 ( 1 )
## 7 ( 1 )
## 8 ( 1 )
## 9 ( 1 )
## 10 ( 1 )
## 11 ( 1 )
## 12 ( 1 )
## 13 ( 1 )
## 14 ( 1 ) *
## 15 ( 1 ) *par(mar = c(4.5, 4.5, 1, 1), mfrow = c(1, 1))
plot(summary(fwd)$cp, col = "blue", type = "l", xlab = "Model size",
ylab = "Cp")
lines(summary(bwd)$cp, col = "red")
lines(summary(exh)$cp, col = "orange")
# They haven't always selected identical models along the
# path, though in this instance they both found the model
# that minimised the overall Cp (confirmed through an
# exhaustive search).- Use the plot method associated with
regsubsetsobjects to visualise the BIC for the various models identified.
par(mfrow = c(3, 1))
plot(fwd, main = "Forward")
plot(bwd, main = "Backward")
plot(exh, main = "Exhaustive")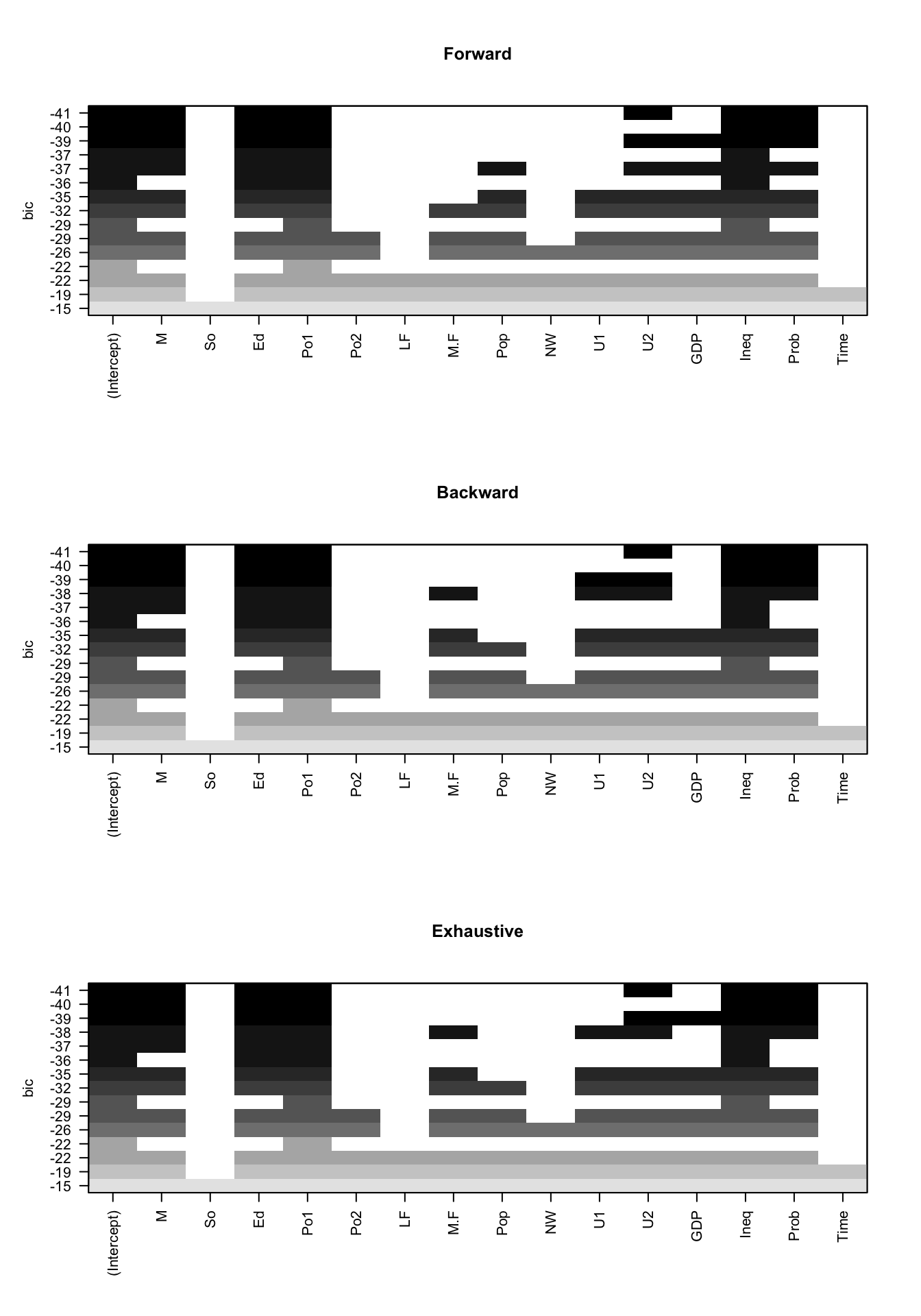
# To have a different criterion on the y-axis in the plots
# specify scale=c('bic', 'Cp', 'adjr2', 'r2') in plot()- The
carpackage offers alternative plot methods to visualise the results from a call toregsubsets(). The code below plots \(C_p\) against dimension for aregsubsetsobject calledexh. Try plotting adjusted \(R^2\) against dimension.
library(car)## Loading required package: carDatapar(mar = c(4.5, 4.5, 1, 1), mfrow = c(1, 1))
subsets(exh, statistic = "cp", legend = FALSE, max.size = 8)## Abbreviation
## M M
## So S
## Ed E
## Po1 P1
## Po2 P2
## LF L
## M.F M.
## Pop Pp
## NW N
## U1 U1
## U2 U2
## GDP G
## Ineq I
## Prob Pr
## Time Tabline(a = 0, b = 1, lty = 2)
subsets(exh, statistic = "adjr2", legend = FALSE, max.size = 10)
## Abbreviation
## M M
## So S
## Ed E
## Po1 P1
## Po2 P2
## LF L
## M.F M.
## Pop Pp
## NW N
## U1 U1
## U2 U2
## GDP G
## Ineq I
## Prob Pr
## Time TBirthweight data
The second example is the birthwt dataset from the MASS package which has data on 189 births at the Baystate Medical Centre, Springfield, Massachusetts during 1986 (Venables and Ripley, 2002) The main variable of interest is low birth weight, a binary response variable low (Hosmer and Lemeshow, 1989). Take the same approach to modelling the full model as in Venables and Ripley (2002), where ptl is reduced to a binary indicator of past history and ftv is reduced to a factor with three levels:
library(MASS)
bwt <- with(birthwt, {
race <- factor(race, labels = c("white", "black", "other"))
ptd <- factor(ptl > 0)
ftv <- factor(ftv)
levels(ftv)[-(1:2)] <- "2+"
data.frame(low = factor(low), age, lwt, race, smoke = (smoke >
0), ptd, ht = (ht > 0), ui = (ui > 0), ftv)
})
options(contrasts = c("contr.treatment", "contr.poly"))
bw.glm1 <- glm(low ~ ., family = binomial, data = bwt)
bw.glm0 <- glm(low ~ 1, family = binomial, data = bwt)
summary(bw.glm1)##
## Call:
## glm(formula = low ~ ., family = binomial, data = bwt)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -1.7038 -0.8068 -0.5008 0.8835 2.2152
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 0.82302 1.24471 0.661 0.50848
## age -0.03723 0.03870 -0.962 0.33602
## lwt -0.01565 0.00708 -2.211 0.02705 *
## raceblack 1.19241 0.53597 2.225 0.02609 *
## raceother 0.74069 0.46174 1.604 0.10869
## smokeTRUE 0.75553 0.42502 1.778 0.07546 .
## ptdTRUE 1.34376 0.48062 2.796 0.00518 **
## htTRUE 1.91317 0.72074 2.654 0.00794 **
## uiTRUE 0.68019 0.46434 1.465 0.14296
## ftv1 -0.43638 0.47939 -0.910 0.36268
## ftv2+ 0.17901 0.45638 0.392 0.69488
## ---
## Signif. codes:
## 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 234.67 on 188 degrees of freedom
## Residual deviance: 195.48 on 178 degrees of freedom
## AIC: 217.48
##
## Number of Fisher Scoring iterations: 4Exercises
- Use forward and backward stepwise to identify potential models. Do you get the same model using both approaches?
step(bw.glm1)## Start: AIC=217.48
## low ~ age + lwt + race + smoke + ptd + ht + ui + ftv
##
## Df Deviance AIC
## - ftv 2 196.83 214.83
## - age 1 196.42 216.42
## <none> 195.48 217.48
## - ui 1 197.59 217.59
## - smoke 1 198.67 218.67
## - race 2 201.23 219.23
## - lwt 1 200.95 220.95
## - ht 1 202.93 222.93
## - ptd 1 203.58 223.58
##
## Step: AIC=214.83
## low ~ age + lwt + race + smoke + ptd + ht + ui
##
## Df Deviance AIC
## - age 1 197.85 213.85
## <none> 196.83 214.83
## - ui 1 199.15 215.15
## - race 2 203.24 217.24
## - smoke 1 201.25 217.25
## - lwt 1 201.83 217.83
## - ptd 1 203.95 219.95
## - ht 1 204.01 220.01
##
## Step: AIC=213.85
## low ~ lwt + race + smoke + ptd + ht + ui
##
## Df Deviance AIC
## <none> 197.85 213.85
## - ui 1 200.48 214.48
## - smoke 1 202.57 216.57
## - race 2 205.47 217.47
## - lwt 1 203.82 217.82
## - ptd 1 204.22 218.22
## - ht 1 205.16 219.16##
## Call: glm(formula = low ~ lwt + race + smoke + ptd + ht + ui, family = binomial,
## data = bwt)
##
## Coefficients:
## (Intercept) lwt raceblack raceother
## -0.12533 -0.01592 1.30086 0.85441
## smokeTRUE ptdTRUE htTRUE uiTRUE
## 0.86658 1.12886 1.86690 0.75065
##
## Degrees of Freedom: 188 Total (i.e. Null); 181 Residual
## Null Deviance: 234.7
## Residual Deviance: 197.9 AIC: 213.9step(bw.glm0, scope = list(upper = bw.glm1, lower = bw.glm0),
method = "forward")## Start: AIC=236.67
## low ~ 1
##
## Df Deviance AIC
## + ptd 1 221.90 225.90
## + lwt 1 228.69 232.69
## + ui 1 229.60 233.60
## + smoke 1 229.81 233.81
## + ht 1 230.65 234.65
## + race 2 229.66 235.66
## + age 1 231.91 235.91
## <none> 234.67 236.67
## + ftv 2 232.09 238.09
##
## Step: AIC=225.9
## low ~ ptd
##
## Df Deviance AIC
## + age 1 217.30 223.30
## + lwt 1 217.50 223.50
## + ht 1 217.66 223.66
## + race 2 217.02 225.02
## + ui 1 219.12 225.12
## + smoke 1 219.33 225.33
## + ftv 2 217.88 225.88
## <none> 221.90 225.90
## - ptd 1 234.67 236.67
##
## Step: AIC=223.3
## low ~ ptd + age
##
## Df Deviance AIC
## + ht 1 213.12 221.12
## + lwt 1 214.31 222.31
## + smoke 1 215.04 223.04
## + ui 1 215.13 223.13
## <none> 217.30 223.30
## + race 2 213.97 223.97
## + ftv 2 214.63 224.63
## - age 1 221.90 225.90
## - ptd 1 231.91 235.91
##
## Step: AIC=221.12
## low ~ ptd + age + ht
##
## Df Deviance AIC
## + lwt 1 207.43 217.43
## + ui 1 210.13 220.13
## + smoke 1 210.89 220.89
## <none> 213.12 221.12
## + race 2 210.06 222.06
## + ftv 2 210.38 222.38
## - ht 1 217.30 223.30
## - age 1 217.66 223.66
## - ptd 1 227.93 233.93
##
## Step: AIC=217.43
## low ~ ptd + age + ht + lwt
##
## Df Deviance AIC
## + ui 1 205.15 217.15
## + smoke 1 205.39 217.39
## <none> 207.43 217.43
## + race 2 203.77 217.77
## - age 1 210.12 218.12
## + ftv 2 204.33 218.33
## - lwt 1 213.12 221.12
## - ht 1 214.31 222.31
## - ptd 1 219.88 227.88
##
## Step: AIC=217.15
## low ~ ptd + age + ht + lwt + ui
##
## Df Deviance AIC
## <none> 205.15 217.15
## + smoke 1 203.24 217.24
## + race 2 201.25 217.25
## - ui 1 207.43 217.43
## - age 1 207.51 217.51
## + ftv 2 202.41 218.41
## - lwt 1 210.13 220.13
## - ht 1 212.70 222.70
## - ptd 1 215.48 225.48##
## Call: glm(formula = low ~ ptd + age + ht + lwt + ui, family = binomial,
## data = bwt)
##
## Coefficients:
## (Intercept) ptdTRUE age htTRUE
## 1.74560 1.42123 -0.05331 1.90580
## lwt uiTRUE
## -0.01437 0.69193
##
## Degrees of Freedom: 188 Total (i.e. Null); 183 Residual
## Null Deviance: 234.7
## Residual Deviance: 205.2 AIC: 217.2- Use the
bestglm()function from thebestglmpackage to identify the top 10 models according to the AIC.
library(bestglm)
# the Xy matrix needs y as the right-most variable:
bwt.Xy = bwt[, -1]
bwt.Xy = cbind(bwt.Xy, low = bwt$low)
bglm.AIC = bestglm(Xy = bwt.Xy, family = binomial, IC = "AIC",
TopModels = 10)## Morgan-Tatar search since family is non-gaussian.
## Note: factors present with more than 2 levels.bglm.AIC$BestModels## age lwt race smoke ptd ht ui ftv Criterion
## 1 FALSE TRUE TRUE TRUE TRUE TRUE TRUE FALSE 211.8516
## 2 FALSE TRUE TRUE TRUE TRUE TRUE FALSE FALSE 212.4823
## 3 TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE 212.8337
## 4 TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE 213.1514
## 5 FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 214.4174
## 6 FALSE TRUE TRUE FALSE TRUE TRUE TRUE FALSE 214.5666
## 7 FALSE TRUE TRUE TRUE TRUE TRUE FALSE TRUE 214.7772
## 8 TRUE TRUE FALSE FALSE TRUE TRUE TRUE FALSE 215.1535
## 9 TRUE TRUE FALSE TRUE TRUE TRUE TRUE FALSE 215.2403
## 10 TRUE TRUE TRUE FALSE TRUE TRUE TRUE FALSE 215.2470summary(bglm.AIC$BestModel)##
## Call:
## glm(formula = y ~ ., family = family, data = Xi, weights = weights)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -1.7308 -0.7841 -0.5144 0.9539 2.1980
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -0.125326 0.967561 -0.130 0.89694
## lwt -0.015918 0.006954 -2.289 0.02207 *
## raceblack 1.300856 0.528484 2.461 0.01384 *
## raceother 0.854414 0.440907 1.938 0.05264 .
## smokeTRUE 0.866582 0.404469 2.143 0.03215 *
## ptdTRUE 1.128857 0.450388 2.506 0.01220 *
## htTRUE 1.866895 0.707373 2.639 0.00831 **
## uiTRUE 0.750649 0.458815 1.636 0.10183
## ---
## Signif. codes:
## 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 234.67 on 188 degrees of freedom
## Residual deviance: 197.85 on 181 degrees of freedom
## AIC: 213.85
##
## Number of Fisher Scoring iterations: 4Rock-wallabies: cross validation
Use the cross-validation argument in bestglm to identify the best rock-wallaby model using cross-validation.
data("wallabies", package = "mplot")
names(wallabies)## [1] "rw" "edible" "inedible" "canopy" "distance"
## [6] "shelter" "lat" "long"wdat = data.frame(subset(wallabies, select = -c(lat, long)),
EaD = wallabies$edible * wallabies$distance, EaS = wallabies$edible *
wallabies$shelter, DaS = wallabies$distance * wallabies$shelter)
X = subset(wdat, select = -rw)
y = wdat$rw
Xy = as.data.frame(cbind(X, y))rwglm.CV = bestglm(Xy = Xy, family = binomial, IC = "CV")## Morgan-Tatar search since family is non-gaussian.summary(rwglm.CV$BestModel)##
## Call:
## glm(formula = y ~ ., family = family, data = data.frame(Xy[,
## c(bestset[-1], FALSE), drop = FALSE], y = y))
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -2.1177 -1.1279 0.6382 1.0923 1.3685
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -0.43867 0.22213 -1.975 0.0483 *
## edible 0.06422 0.01555 4.130 3.62e-05 ***
## ---
## Signif. codes:
## 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 272.12 on 199 degrees of freedom
## Residual deviance: 248.45 on 198 degrees of freedom
## AIC: 252.45
##
## Number of Fisher Scoring iterations: 4Advanced: Stagewise selection
The basic idea behind stagewise modelling is:
- Standardise the predictors, \(\tilde{x}_{ij} = (x_{ij} - \bar{x}_j)/\text{sd}(x_j)\), and centre the responses, \(\tilde{y}_i = y_i - \bar{y}\).
- Initialise \(\hat{\boldsymbol{\mu}} = (0,\ldots,0)'\), \(\mathbf{r} = \tilde{\mathbf{y}}\), and \(\hat{\beta}_1 = \ldots = \hat{\beta}_p = 0\).
-
For
i in large_number:- Find the predictor \(\tilde{x}_j\) most correlated with \(\mathbf{r}\) (the residual vector). This is also the same as picking the variable that would result in the biggest drop in RSS.
-
Set \(\delta = \epsilon\times\text{sign}(\text{cor}(\tilde{\mathbf{x}}_j,\mathbf{r}))\) and update the parameters:
- \(\hat{\beta}_j = \hat{\beta}_j + \delta\)
- \(\hat{\boldsymbol{\mu}} = \hat{\boldsymbol{\mu}} + \delta\tilde{\mathbf{x}}_j\)
- \(\mathbf{r} = \mathbf{r} - \delta\tilde{\mathbf{x}}_j\)
Exercise
Implement the stagewise procedure for the artificial example introduced in the lecture. Hint: the core of the function will look like this:
data("artificialeg",package="mplot")
y = artificialeg$y-mean(artificialeg$y)
X = artificialeg[,-10]
y = # mean zero predictors
M = # standardised predictor matrix
lots = 20000
beta = matrix(0, ncol=ncol(M), nrow=lots)
r = y
eps = 0.001
for (i in 2:lots){
co = cor(M,r)
j =
delta =
b = beta[i-1,]
b[j] =
beta[i,] = b
r =
}- Visualise the path of the estimated coefficients against step number. Hint: use
matplot(). - Visualise the path of the estimated coefficients against the sum of the absolute values of the coefficients.
data("artificialeg", package = "mplot")
y = artificialeg$y - mean(artificialeg$y)
X = artificialeg[, -10]
s = apply(X, 2, sd)
M = scale(X)
lots = 20000
beta = matrix(0, ncol = ncol(M), nrow = lots)
r = y
eps = 0.001
for (i in 2:lots) {
co = cor(M, r)
j = which.max(abs(co))
delta = eps * sign(co[j])
beta[i, ] = beta[i - 1, ]
beta[i, j] = beta[i, j] + delta
r = r - delta * M[, j]
}
# visualise the path of the coefficients
par(mar = c(4.5, 4.5, 2, 1))
matplot(beta, type = "l", lty = 1, xlab = "step number", ylab = "beta",
main = "stagewise")
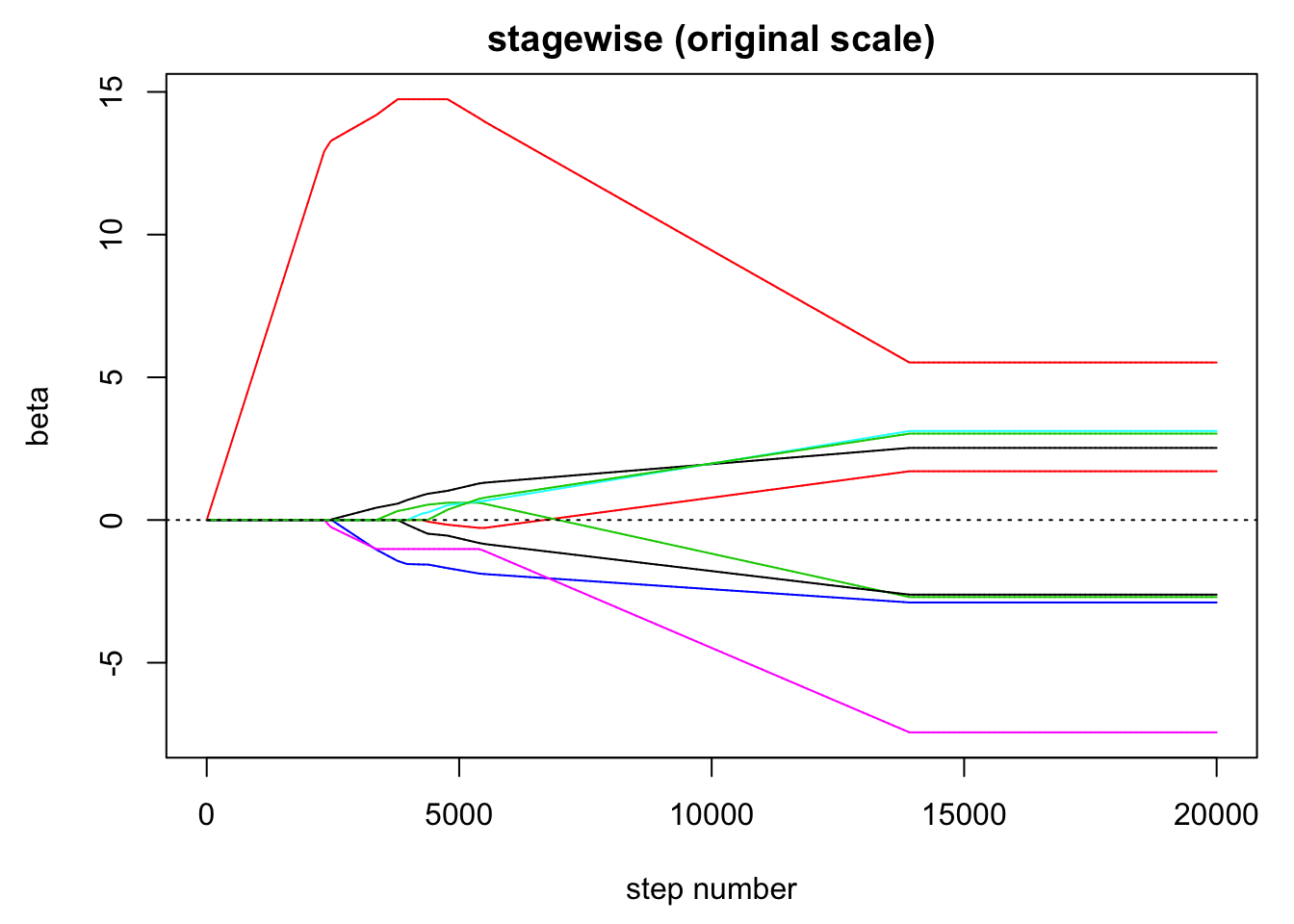
abline(h = 0, lty = 3)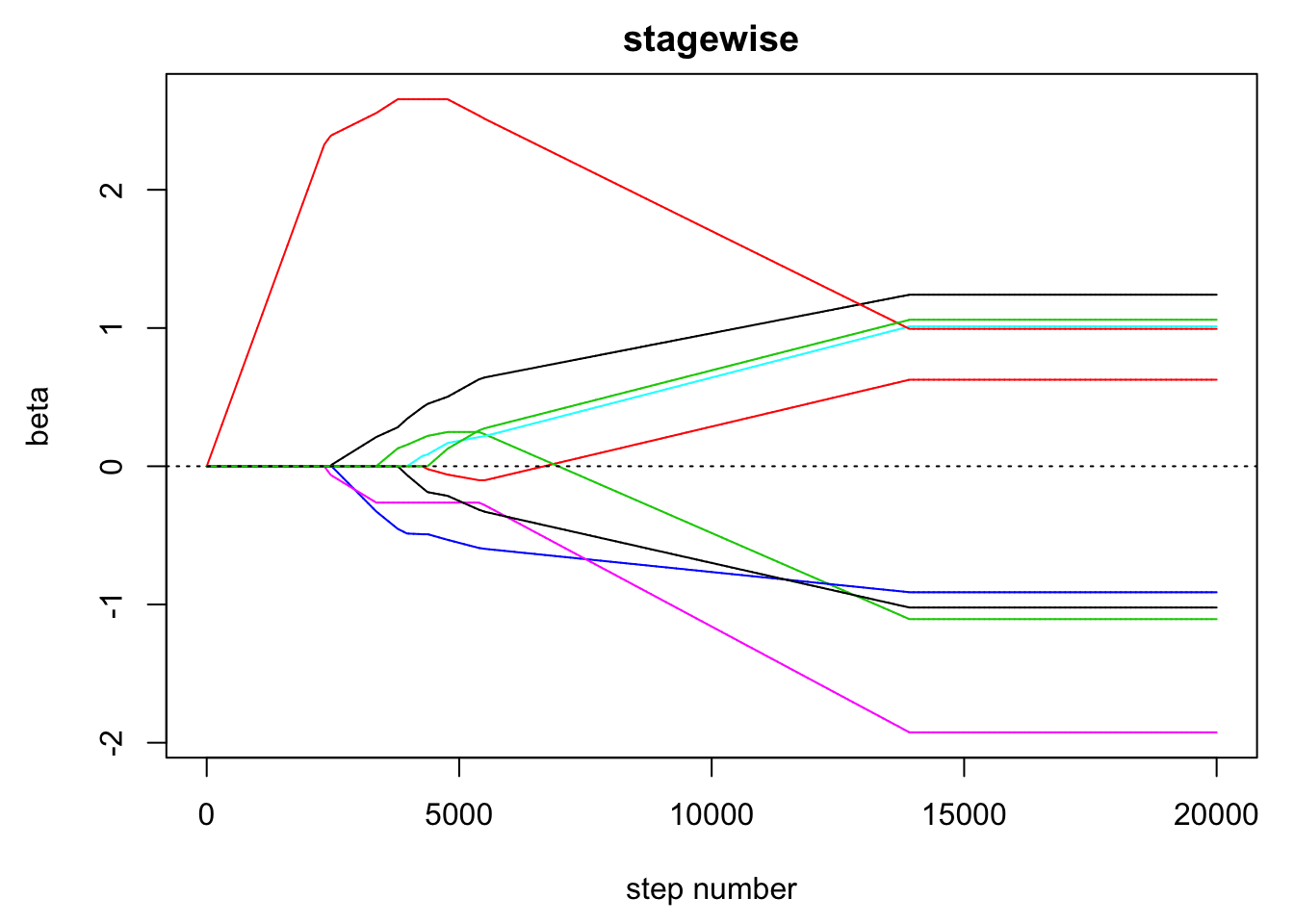
# Sometimes it is useful to visualise the estimated
# coefficients as a function of the sum of the absolute
# values of the coeffs:
matplot(apply(beta, 1, function(x) sum(abs(x))), beta, type = "l",
lty = 1, xlab = "sum abs(beta)", ylab = "beta", main = "stagewise")
abline(h = 0, lty = 3)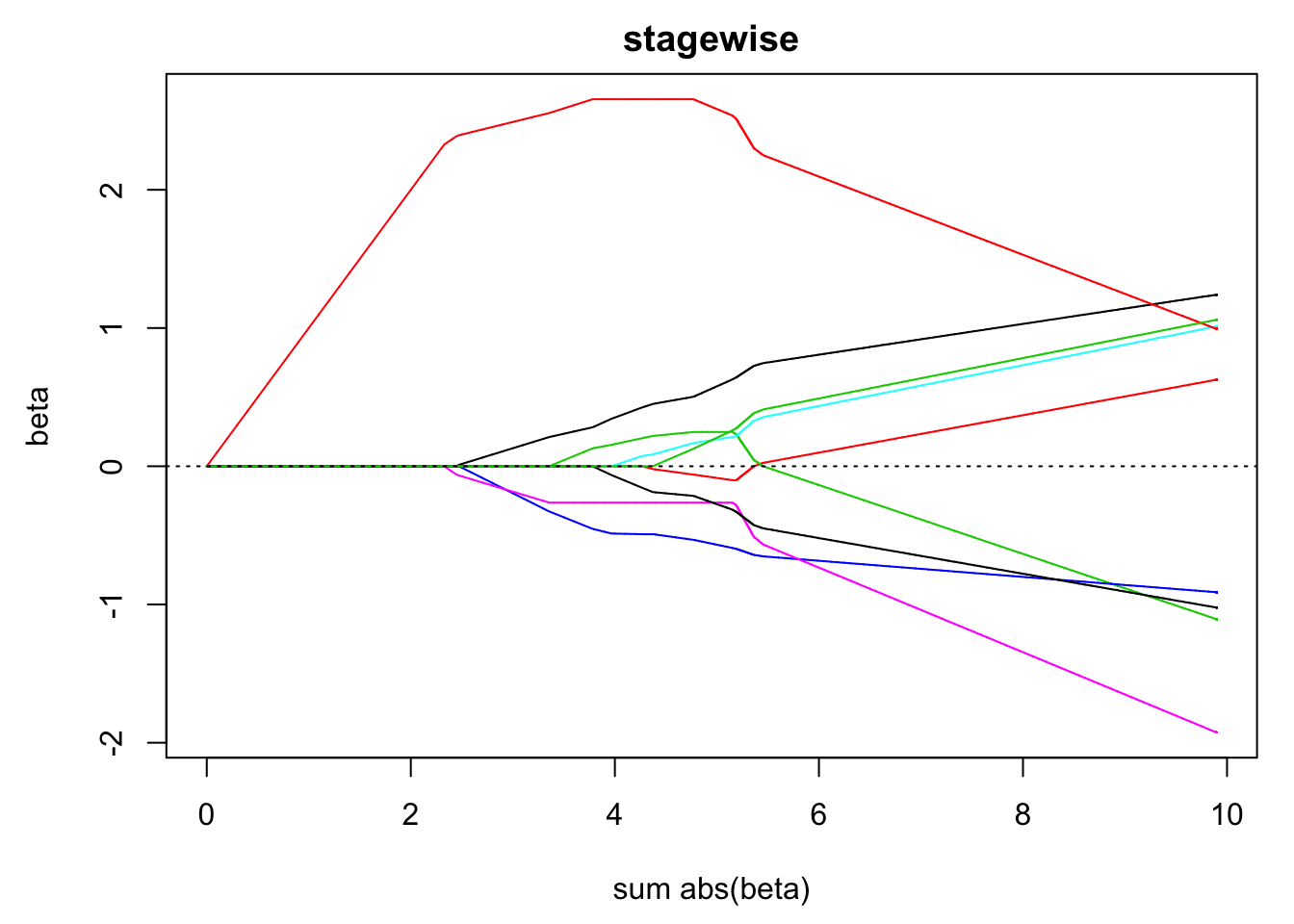
# on the original scale
unscale = scale(beta, center = FALSE, scale = 1/s)
matplot(unscale, type = "l", lty = 1, xlab = "step number", ylab = "beta",
main = "stagewise (original scale)")
abline(h = 0, lty = 3)